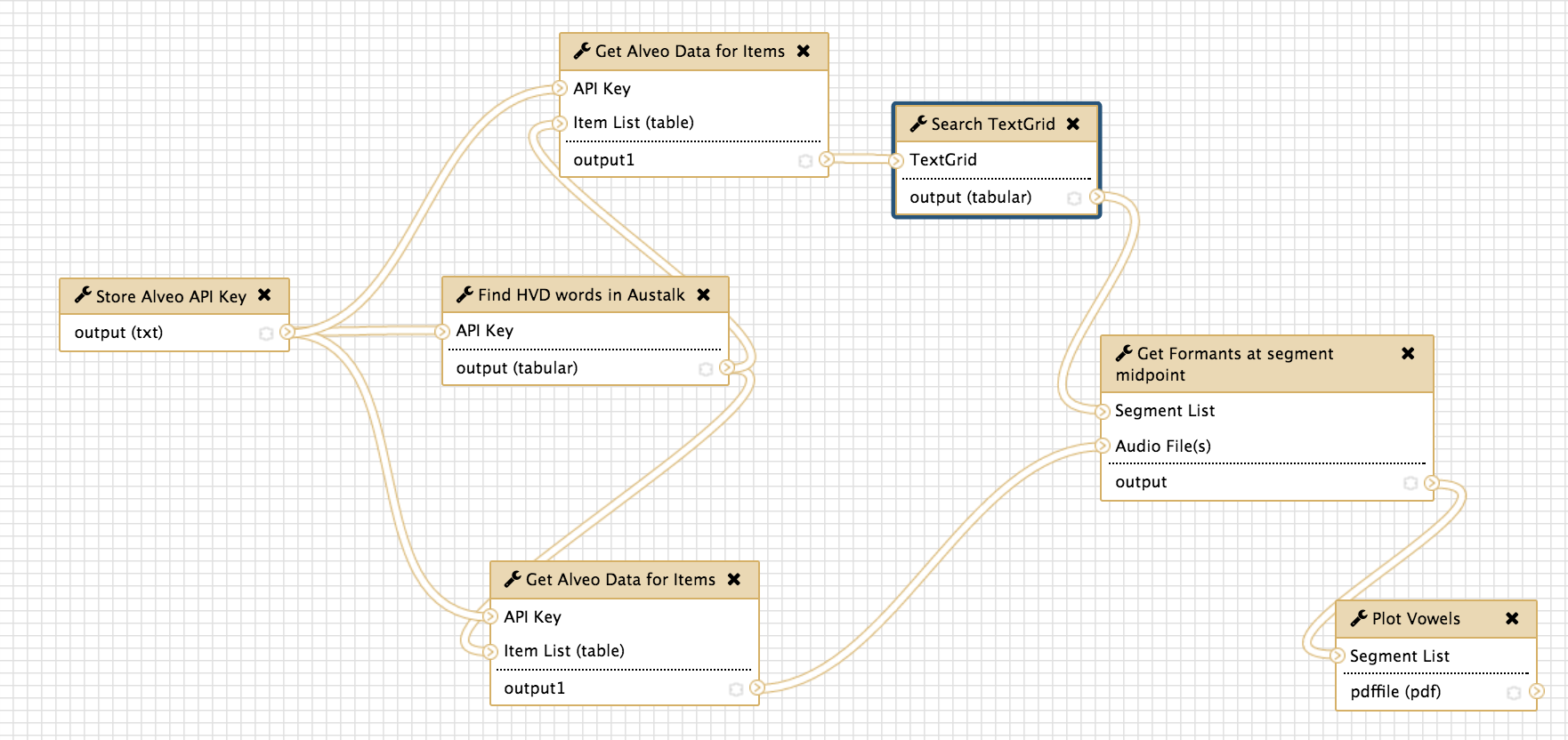
A Galaxy Workflow for Acoustic Phonetic Analysis
I’ve been working for a while now on adapting the Galaxy Workflow engine for use in speech analysis, specifically for acoustic phonetic analysis of vowel sounds. Galaxy is a system used in bioinformatics for constructing workflows to do genetic analysis and other things. As part of the Alveo project we’ve been building tools for doing text and speech analysis for Galaxy. My recent work has been specifically looking at acoustic phonetic analysis with the Emu library with a goal of reproducing some work on children’s vowels that I did with Catherine Watson many years ago.
I’ve just managed to get the first full workflow going. It extracts the hVd monophthongs for a single speaker from the Austalk corpus stored in Alveo, finds vowels in the TextGrid files via the python tgt package, uses the wrassp package from Emu to compute the formants and then plots them with the phonR library. The workflow is shown at the top of this post in the edit view of Galaxy - this shows the individual tools that are used for each step of the process. The end result is a PDF plot generated by R and reproduced here as a PNG image.
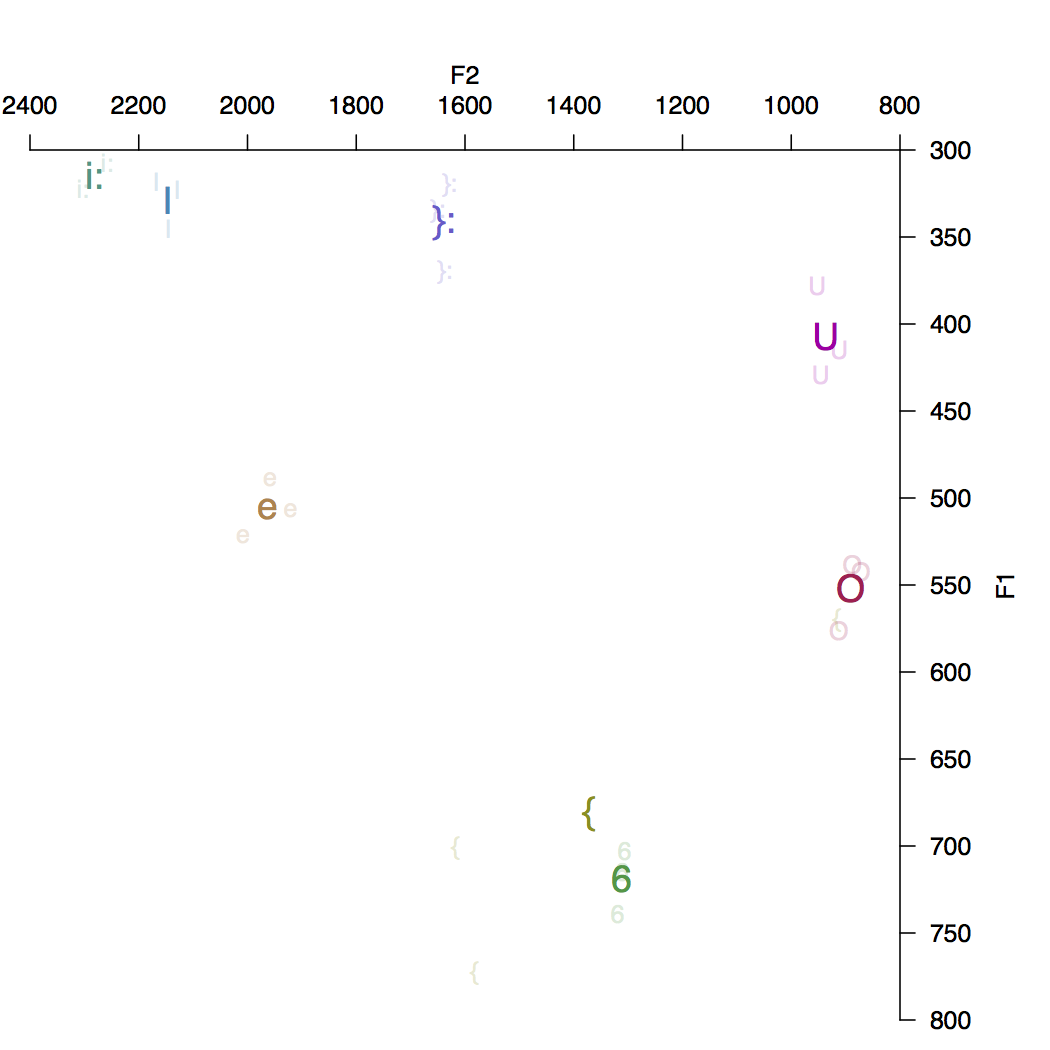
The plot here is for speaker 1_1308 from the Austalk corpus (a 35yr old male from Sydney, born in Adelaide). There is clearly a formant tracking error with the { vowel (had) in one instance (putting it inside the O cluster). Just to prove that it works with any speaker here’s a second example from speaker 1_366 (a 73yr old female from Adelaide); in this case something odd is going on with the i: vowel where F2 is zero for the two instances found.
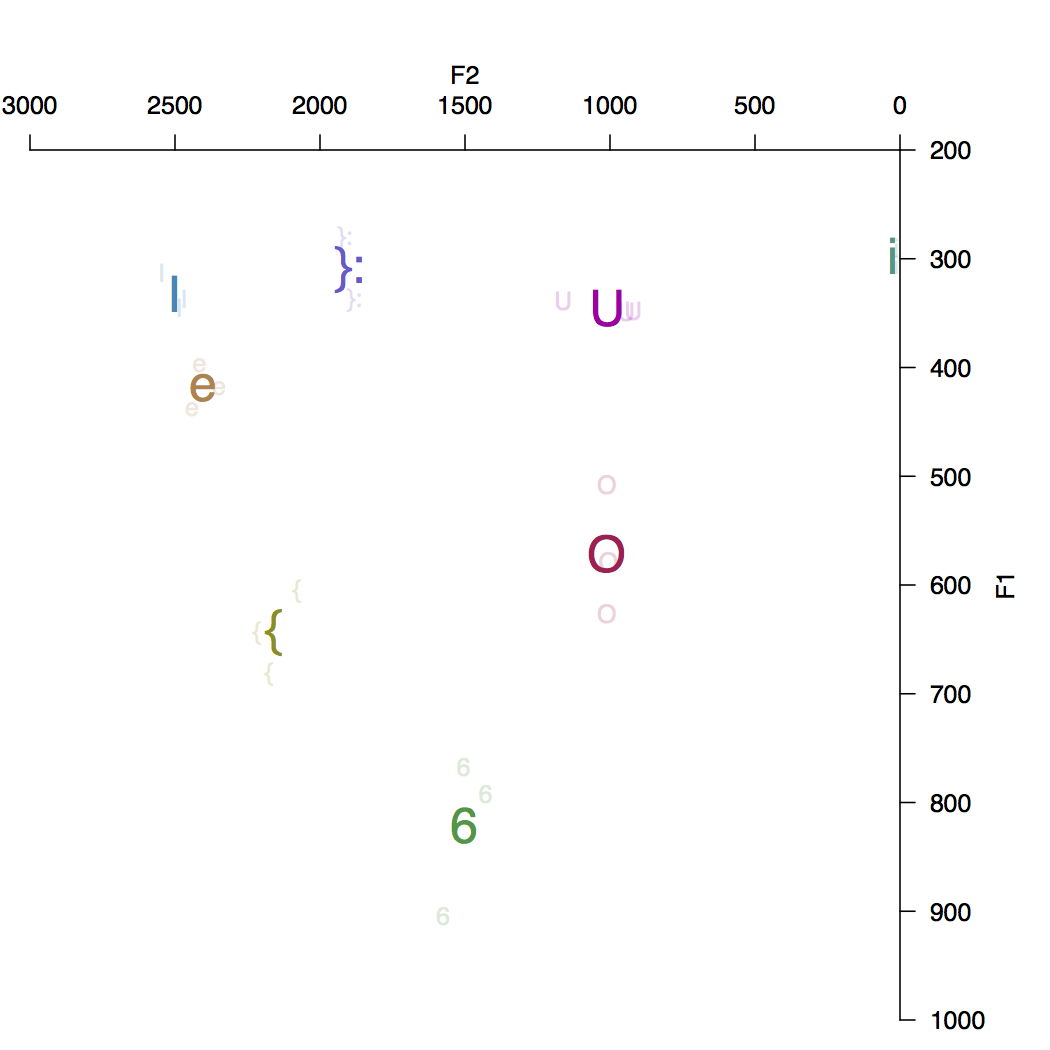
This workflow relies on there being stored TextGrid files for the speaker in question. An obvious next step is to incorporate a forced-alignment tool to derive vowel segment boundaries given the transcription. I’m currently looking at options for doing this with MAUS and/or FAVE.
The hardest part of getting to this point has been dealing with the underlying Galaxy infrastructure. We had an earlier Galaxy installation for the Alveo project but it was based on an older version of Galaxy, which is a rapidly moving project. A significant change has been the use of Conda to manage package dependencies. I’ve been working on writing conda packages for the tools that I’ve used here so that I could write Galaxy tools that made use of them. This has been helped a lot by the Bioconda repository run by Björn Grüning which automates building of these packages and makes them available via the bioconda channel (bioconda is the home of many biotechnology packages used in Galaxy, we’ve been invited to submit speech and NLP packages there since we’re working with Galaxy too). Once these dependencies are in place, I can write tools that use them and have them installed in a Galaxy instance. I’ve been using Docker to build a Galaxy flavour for Alveo which installs a set of tools useful for speech and text analysis. These include some of the tools we developed earlier in the Alveo project and add some new tools for speech analysis.
A major issue when writing tools for Galaxy is the level of granularity to pitch them at. For example, it would be possible to write a single tool script to carry out this entire workflow, but that then removes the possibility that parts of the workflow could be re-used for other tasks. For example, I also want to generate a plot of the durations of the vowel segments, so I need access to the intermediate result that is the output of the query step. One choice I made was to build a single tool to accept a list of vowel segments and run the wrassp formant tracker over them and then select the value at the vowel midpoint. This could be decomposed into smaller steps that would then allow the intermediate formant tracks to be re-used for other tasks. I’m still considering the best way to approach this.
I now have a version of my Galaxy server running with these tools installed. It is not quite ready for production but at some point it will be accessible at galaxy.alveo.edu.au to replace our older server. At that point I will be able to share this workflow with others so that you can run it yourself on other data from Austalk.